Constructing Pedigrees in R
I was looking for a good way to draw pedigrees using software, without having to fuss drawing them “by hand” using a drawing program. Lo and behold, I came across a few different packages in R that allow me to do just that!
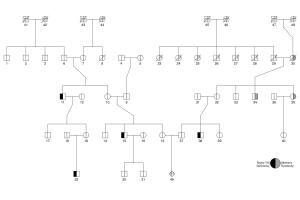
Using the kinship2 package, I was quickly able to produce a nice pedigree for a family with two genetic conditions: factor VIII deficiency (haemophilia) and myotonic dystrophy. Factor VIII deficiency is a classic example of a sex-linked recessive trait, and myotonic dystrophy follows autosomal dominant inheritance. In this pedigree, I show the status of affected and unaffected indiviudals (not carriers), using the following bits of code in R.
First, let’s install (if needed) and load the kinship2 package:
if (!require(kinship2)) {install.packages("kinship2"); library(kinship2)}
Now we need to code several variables which will correspond to all of the individuals in the pedigree. First, we need individual ID numbers:
ids <- 1:49
Second, we need to identify the father and mother of each individual using the individual ID numbers above:
dad <- c(41,41,41,NA,NA,41,43,43,4,6,6,NA,9,9,9,NA,11,11,NA,15,
15,19,45,45,45,45,45,45,45,47,NA,29,29,29,29,NA,31,31,31,36,
NA,NA,NA,NA,NA,NA,NA,NA,37)
mom <- c(42,42,42,NA,NA,42,44,44,5,7,7,NA,10,10,10,NA,12,12,NA,16,
16,18,46,46,46,46,46,46,46,48,NA,30,30,30,30,NA,32,32,32,35,
NA,NA,NA,NA,NA,NA,NA,NA,13)
Now we code the sex and status (0=alive, 1=dead) of each individual:
sexes <- c(1,1,1,1,2,1,2,2,1,2,1,2,2,1,1,2,1,2,1,1,1,1,2,2,2,1,1,1,1,
2,1,2,1,1,2,1,1,1,2,2,1,2,1,2,1,2,1,2,3)
dead <- c(0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1,1,1,1,1,1,1,
0,0,0,0,0,0,0,0,0,0,1,1,1,1,1,1,1,1,0)
Here is where we code the affected status of each individual (0=unaffected, 1=affected, NA=unknown). We need to bind these columns together in a matrix (you can have up to 4 traits; one per column):
F8 <- c(0,0,0,0,0,0,0,0,0,0,1,0,0,0,1,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,
0,0,0,0,0,0,0,1,0,0,NA,NA,NA,NA,NA,NA,NA,NA,NA)
MD <- c(0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,NA,NA,NA,NA,NA,NA,NA,1,
0,0,0,1,1,0,0,0,0,0,NA,NA,NA,NA,NA,NA,NA,NA,NA)
affect <- as.matrix(cbind(F8, MD))
And now we create the pedigree object and plot it:
family <- pedigree(id=ids, dadid=dad, momid=mom,
affected=affect, sex=sexes, status=dead)
plot.pedigree(family)
pedigree.legend(family, location="bottomright",
labels=list("Factor VIII\nDeficiency", "Myotonic\nDystrophy"),
cex=0.8, radius=0.3)
That’s the basics! Of course, with a large pedigree I expect this becomes a bit unruly, but for smaller examples like this one it’s relatively easy to put together.